The human body runs on its own biological computer with DNA as its code. At compile time, DNA synthesizes from its original form into mRNA, converting genetic information into proteins. mRNA is code for proteins synthesized at the ribosome. Since DNA and mRNA occur in sequences, and AI models are good at modeling sequences, AI can play a role in learning about our coding (e.g. our DNA) and that of other organisms.
Enter Large Language Models (LLMs), a class of AI architectures prime for studying sequences. Since proteins, DNA sequences, and their encodings are all mathematical objects (or can be treated as such) from AI’s perspective, we can directly apply tools from Natural Language Processing, a field broadly referring to the application of artificial intelligence to language data, in bioinformatics contexts. In other words, a signal processing problem, like deciphering voices in a noisy room can be seen as analogous to a bioinformatics problem like unraveling the multiple sequences in a meta genome. Moreover, an AI that can predict what a person might say next (made possible via a transformer architecture–covered below–trained on a corpus of text) can also predict how a virus will evolve next if trained on a bioinformatics dataset.
Several groups have jumped at the opportunity to use AI language systems, namely LLMs for the study of human biology. Recent advances in AI and LLMs usemassive text datasets to better understand human language and gain insight into context and sequences of text. But these advances aren’t limited to human language.
Recent applications of LLMs let AI learn the languages of biology and chemistry as well. By making it easier to train massive neural networks on biomolecular data, NVIDIA BioNeMO, for instance, helps researchers discover new patterns and insights in biological sequences — insights that can connect to biological properties, functions, and human health conditions.
Let’s take a look at how this is possible.
First, what is a LLM? LLMs are machine learning algorithms that use enormous text datasets, hence the models are LARGE– often trained on hundreds of gigabytes of text data to recognize, predict, and generate human language on the basis of predicting the next word in a sequence. OpenAI’s GPT-3 (trained on 175 billion parameters) is a popular example.
We cannot understand LLMs without understanding how the technology came about. Before we go further into how LLMs are used, let’s go on a tangent and cover the history of their predecessor architectures used for natural language processing to understand how and why LLMs came about.
NLP History and the birth of the transformer
The basic feed-forward neural network isn’t designed to keep track of sequential data. It maps each individual input to an output, which is helpful for tasks like classifying images, but fails on text due to its sequential nature. To process text, we must take into account sequences and the relationships between words and sentences.
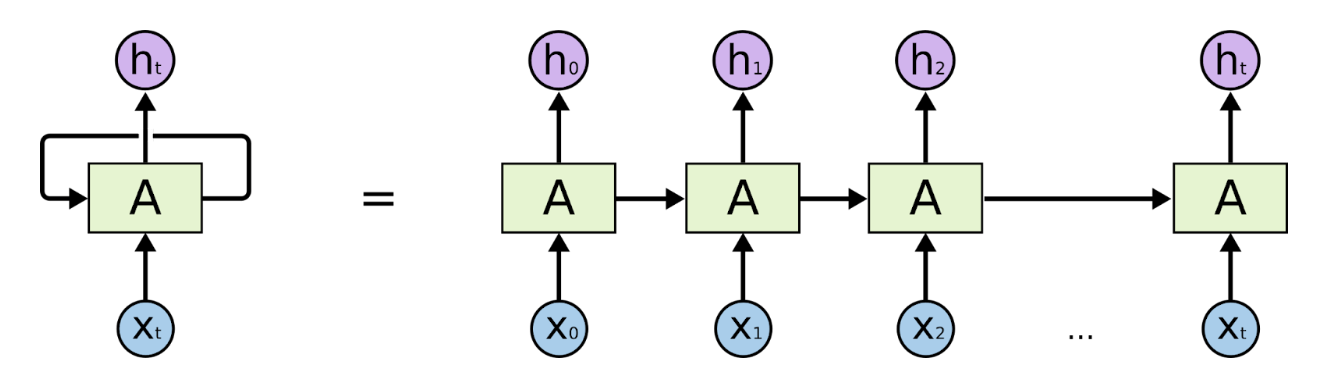
Unrolled RNN Architecture (Source: https://colah.github.io/posts/2015-08-Understanding-LSTMs/)
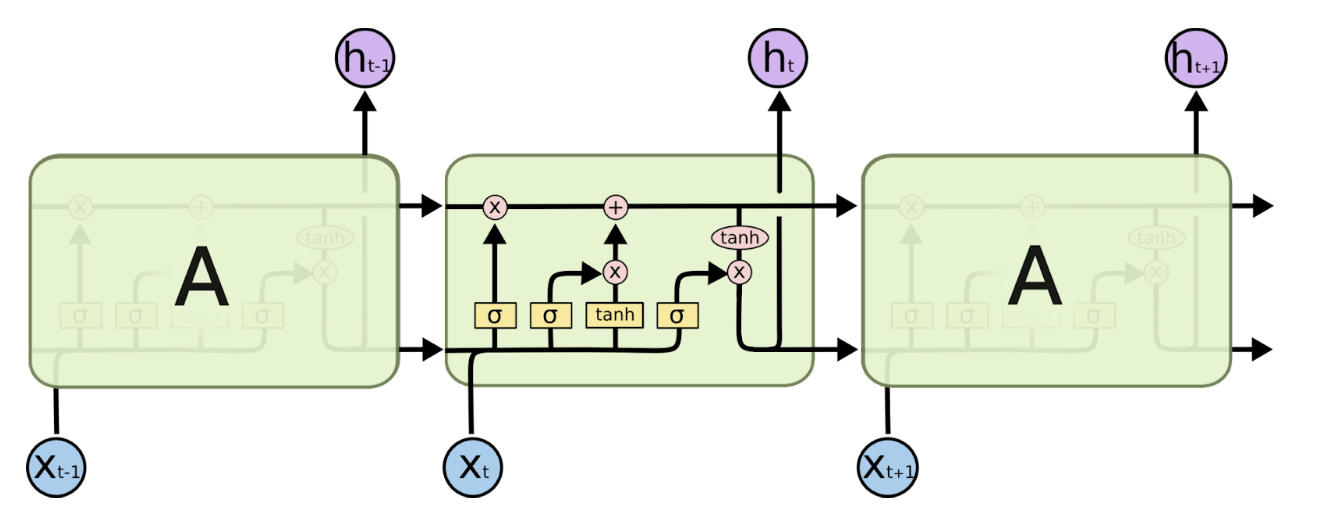
LSTM Architecture (Source: https://colah.github.io/posts/2015-08-Understanding-LSTMs/)
Before transformers were introduced in 2017), Recurrent Neural Networks (RNN) and Long Short-Term Memory networks (LSTM) were popularly used for natural language processing. An RNN processes words by taking the first word and feeding back the result into the layer that processes the next word to keep track of the entire sentence– hence recurrent. They worked well but were slow and could not take advantage of parallel computing hardware or graphics processing units (GPUs). They also exhibited something called “the vanishing gradient problem,” where, as they got deeper into a text excerpt, the early words of the sequence gradually faded, which was problematic for long sequences of text (or DNA in our case). Finally, they could only capture the relations between a given word and those that preceded it (whereas in reality, the words following it also affect meaning). After RNNs came Long Short-Term Memory Networks (LSTMs). They solved the vanishing gradient problem and were able to handle longer text sequences, but were even slower than RNNs at training and still could not support parallel computation.
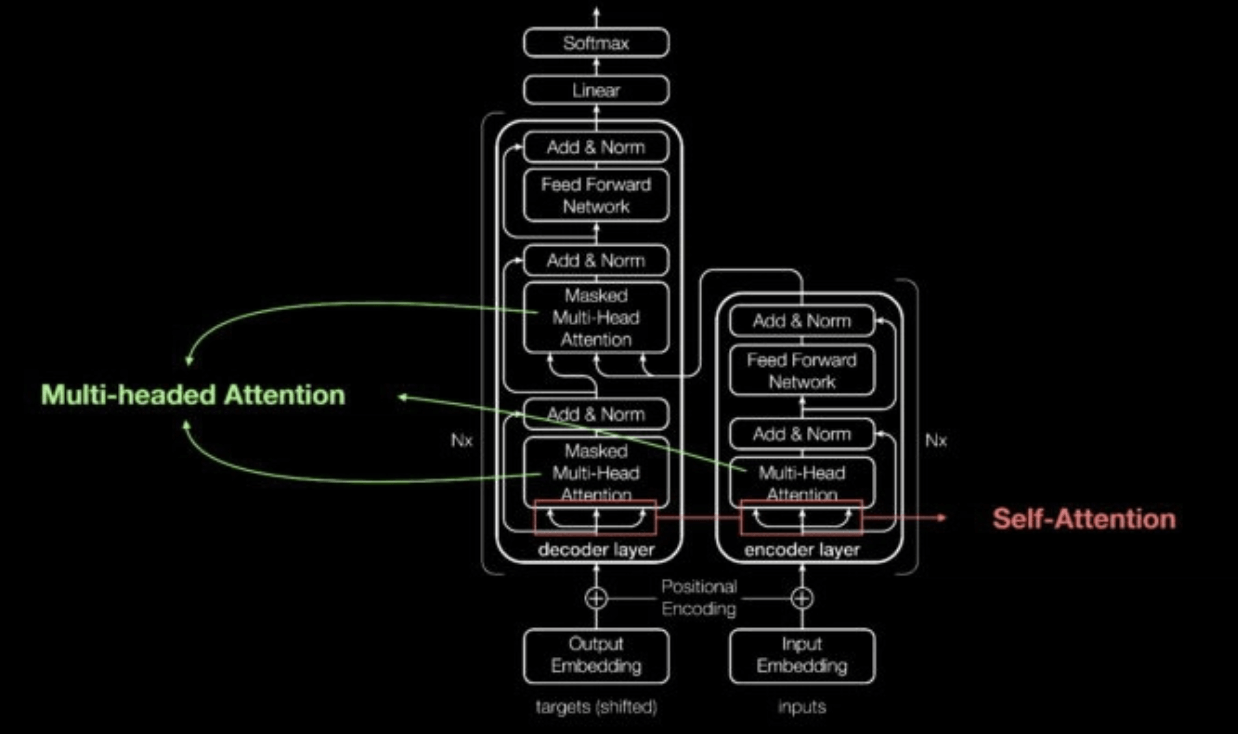
Transformer Self Attention Mechanism Diagram (Link: https://blogs.nvidia.com/blog/2022/03/25/what-is-a-transformer-model/)
Enter transformers in 2017. Transformers can learn context in sequential data. What does this mean? Well, transformer networks make two key contributions: they make it possible to process entire sequences in parallel (which speeds up deep learning), and most importantly, they introduce attention mechanisms, which make it possible to track relationships between words across long text sequences in both directions. This information is called context. [There is ample information online for those looking to learn more about transformers (e.g. Huggingface course and NVIDIA: What are transformer models?)
LLMs are massive transformers and have been used to generate poems, compose music, build websites, write papers, and even code. They can be used to generate text in all forms and are often characterized by their emergent properties, which are still being studied and explored.
Back to biology
So with this immense power to study and manipulate text with LLM neural networks, we turn to a different kind of sequence, DNA. LLMs are already making massive advances in protein and chemical structure study. Applications in drug discovery are underway.
So what’s next?
As the scale of LLMs grows (some are now being trained on upwards of 500 billion parameters), so too can their capabilities expand. The sheer size of the networks alleviates the need to train on custom, smaller datasets for specific tasks and allows scientists to scale up their models and capture information about molecular structure and more.
I am personally working with a group interested in the plankton metagenome and the role LLMs can play in unraveling the metagenome into phenotypes. A cubic mile of ocean water contains billions (if not more) of planktonic organisms. By routinely sampling ocean water, we can collect information and map the metagenome of these organisms. Taken together with metadata such as temperature, acidity, depth, and water pressure, and mapped to a time series, this data reveals changes in the oceanic biome due to changes in climate. By training new foundation models on the extensive metagenome sequences from our samples, we will be able to map changes in ecosystem conditions and climate to changes in the phenotypic manifestations of the animals that live there. Large Language Model (LLM) architectures enable us to take these data-rich genomic sequences and extract information relevant to phenotypes.
I’m even more excited about the potential of LLMs trained on human biochemical sequences. One can imagine replacing the massive repositories of unstructured text data LLMs are currently trained on with massive repositories of unstructured biochemical sequences. An LLM like this might be able to synthesize genomes de novo just as current LLMs are able to synthesize text. The viability of these genomes could be evaluated similarly to how semantic feasibility is evaluated now.
The intersection of LLMs and biochemical research, specifically proteins, drug discovery, and genomics research, is rapidly growing and evolving. I’m excited to see where this field goes next and what major breakthroughs are on the other side of the next few years.



myhome
I agree with your point of view, your article has given me a lot of help and benefited me a lot. Thanks. Hope you continue to write such excellent articles.